A nucleotide is made up of three components: Base (e.g.: Adenine (A), Thymine (T)), sugar and phosphate. The symbols for the most important bases are: G Guanine, A Adenine, C Cytosine, T Thymine and U Uracil.
Oligonucleotides are oligomers made up of a few nucleotides from DNA or RNA. In most cases, the nucleotide sequence for many of the applications consists of between 12 and 40 nucleotide units (corresponding to a 12-mer to 40-mer). Oligos are becoming increasingly important in modern medicine and diagnostics and are used, for example for primers for DNA synthesis, primers for the polymerase chain reaction (PCR), probes for Real Time Quantitative PCR and much more. With the CE, the oligos produced can be tested for purity. For example, if a 20-mer was synthesized, the solution would also contain 19 mer failures, 18 mer failures, 17 mer failures and others. Because of the good resolution of the Capillary Gel Electrophoresis (CGE) method, the amount of failure sequences can be detected.
pd(A) 12 -18
The tests were carried out using the parameters described in this reference:
[1] Agilent application note 5988-4303EN: “Oligonucleotide analysis with the Agilent Capillary Electrophoresis System”, October 1, 2001.
- Mode: Capillary Gel Electrophoresis (CGE)
- Electrolyte: BisTris and Boric acid, each 200 mM
- Capillary: PVA, 33 cm total (Agilent G1600-60419)
- Injection: electrokinetic: -10 kV, 10 s
- Detection: direct UV, 260 nm
- Separation: -25 kV, 30°C
- Description: A system suitability test (SST) of 5 A 260 polydesoxyriboadenine 12-18 („pd(A)12-18“) is shown. In the PDF the SST and the determination of a real sample can be seen.
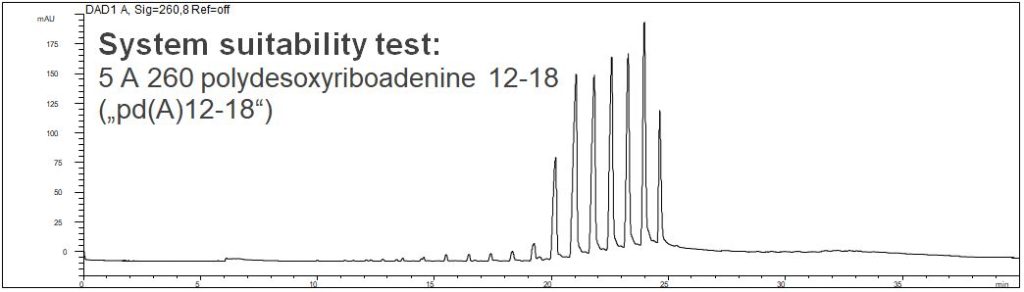
Oligonucleotides: pd(A)12-18
Download PDF: oligo-1.pdf
Oligonucleotide Resolution standard
The tests were carried out using the parameters described in this reference:
Agilent application note 5994-3864EN: “Analysis of Oligonucleotide by Capillary Gel Electrophoresis with the Agilent 7100 CE System”, July 27, 2021.
- Separation: CGE
- Electrolyte: BisTris and Boric acid, each 200 mM
- Gel: PEG 35000
- Capillary: 100 µm ID, PVA coated, 33 cm total (Agilent G1600-60419)
- Detection: direct UV at 260 nm, using the 260 nm filter Agilent: G7100-62700, Remark: The 280 nm filter (part. no. G7100-68750) can be used for this application as well.
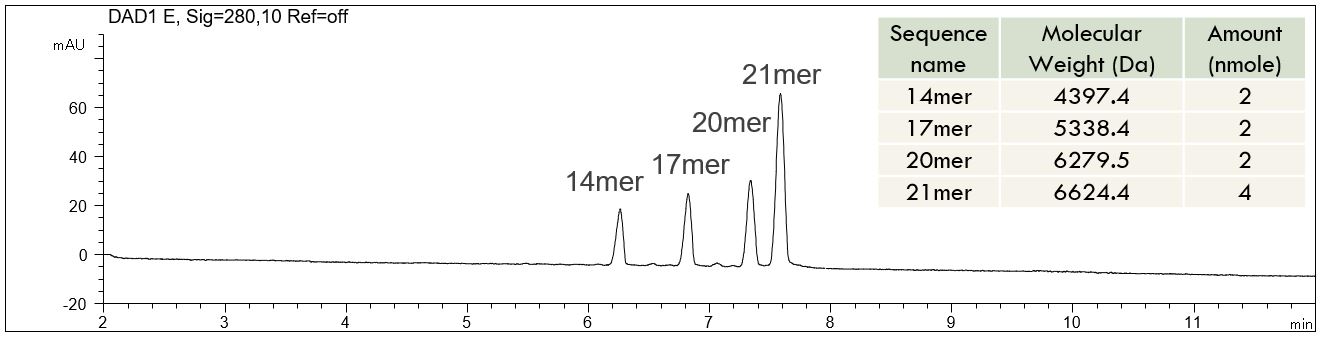
- Description: The RNA resolution standard „Agilent 5190-9028“ was used. For sample preparation 1 ml water was added to the original sample vial and the solution was vortexed for 2 seconds. The PDF (download below the figure) shows the separation of all four oligonucleotides during 8 minutes. The sequence of the oligonucleotides is given in a table in the PDF.
Download: Oligonucleotide resolution standard.pdf
Oligonucleotide pd(A)40-60 Test mix
The tests were carried out using the parameters described in this reference:
Agilent application note 5994-3864EN: “Analysis of Oligonucleotide by Capillary Gel Electrophoresis with the Agilent 7100 CE System”, July 27, 2021.
- Separation: CGE
- Electrolyte: BisTris and Boric acid, each 200 mM
- Gel: PEG 35000
- Capillary: 100 µm ID, PVA coated, 33 cm total (Agilent G1600-60419)
- Detection: direct UV at 260 nm, using the 260 nm filter Agilent: G7100-62700, Remark: The 280 nm filter (part. no. G7100-68750) can be used for this application as well.
- Sample preparation: 0.5 ml water was added to the original sample vial and the solution was vortexed for 2 s. Before the injection the standard solution has been diluted 1:2 with water directly in the micro vial and vortexed for approx. 2 s.
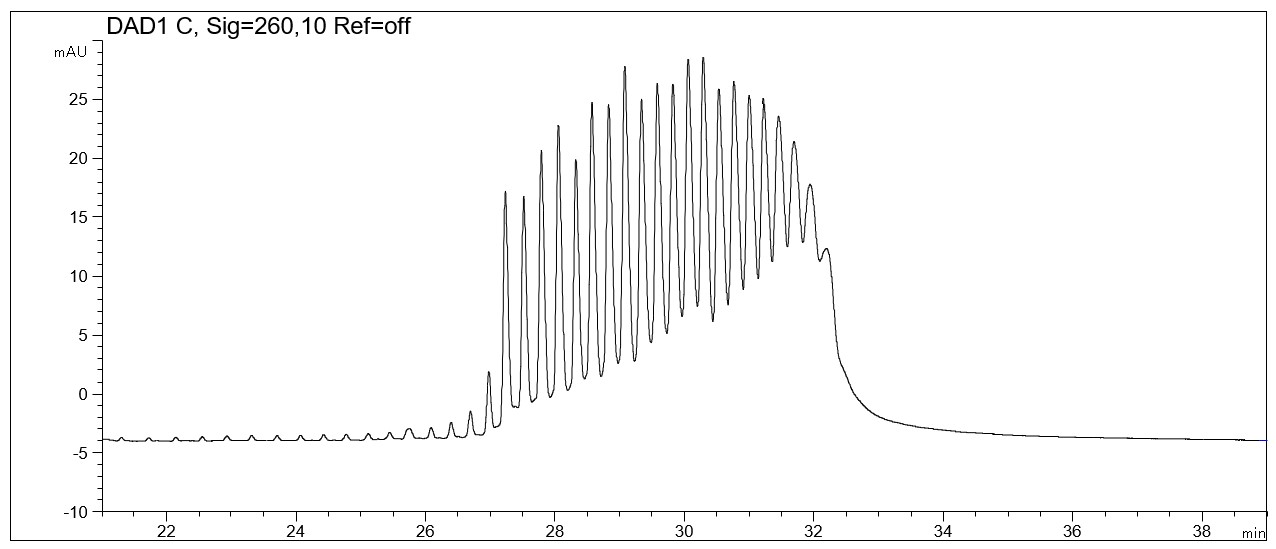
pd(A)40-60 Test mix
Download pdf (133 kB): pdA40-60.pdf
DNA Ladder Standard 15 mer – 40 mer
- Separation: CGE
- Electrolyte: BisTris and Boric acid, each 200 mM
- Gel: PEG 35000
- Capillary: 100 µm ID, PVA coated, 33 cm total (Agilent G1600-60419)
- Detection: direct UV at 260 nm, using the 260 nm filter Agilent: G7100-62700,
- Sample preparation: 1 ml water was added to the original sample vial and the solution was vortexed for 2 s. Before the injection the standard solution has been diluted 1:2 using water directly in the micro vial and vortexed for approx. 2 s.
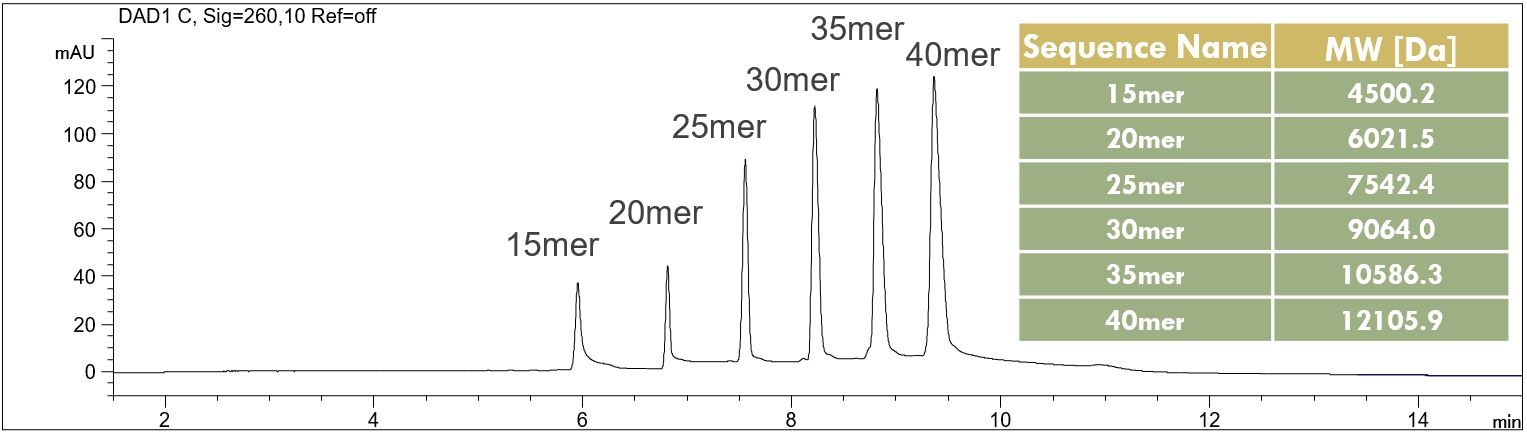
DNA Ladder Standard 15 mer – 40 mer
In the PDF (107 kB) the influence of the capillary length on the resolution of the single Peaks is shown. Download pdf (107 kB): DNA-Ladder-Standard.pdf
The influence of the detector filter on the peak shape and resolution
- Separation: CGE
- Electrolyte: BisTris and Boric acid, each 200 mM
- Gel: PEG 35000
- Capillary: 100 µm ID, PVA coated, 33 cm total (Agilent G1600-60419)
- Detection: direct detection: a) without filter, b) 260 nm filter (Agilent G7100-62700) and c) 280 nm (Agilent G7100- 68750)
- Sample preparation: 1 ml water was added to the original sample vial and the solution was vortexed for 2 s. Before the injection the standard solution has been diluted 1:2 with water directly in the micro vial and vortexed for approx. 2 s.
- Description: The effect of detector filter to the peak sharpness and resolution was investigated. The use of a DAD filter is necessary to obtain sharp peaks and good resolution. In principle, both filters are suitable for the application. However, for the separation of the oligos of interest the 260 nm filter should be preferred.
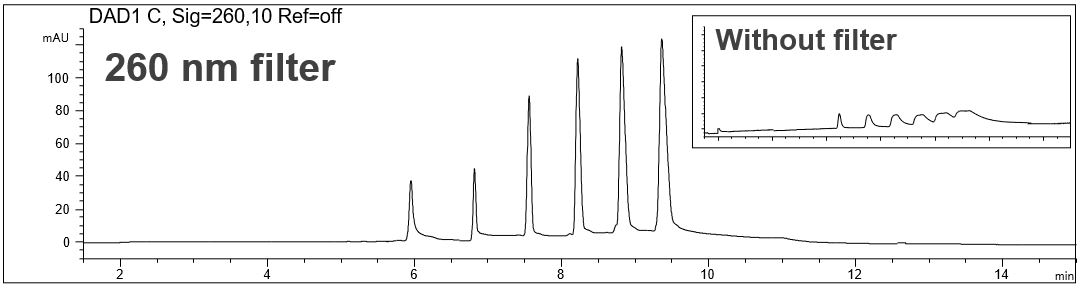
The effect of the detector filter on peak shape
In the PDF (501 KB) the e-grams obtained a) without filter, b) with 260 nm filter and c) with 280 nm filter are shown.
Download pdf (501 kB): oligos-detector-filter